News
12 August 2024
Sinan’s paper on building a haplotype reference panel based on the Genomics England dataset and using it to perform imputation on the UK Biobank has now been published in Nature Genetics!
5 December 2023
Lino has passed his DPhil thesis defence. Congratulations! He will be staying in the group for a little while longer working as a postdoc.
8 November 2023
A new preprint led by Sinan and on which Sile is a co-author has been released on bioRxiv. This paper uses the Genomics England dataset as a haplotype reference panel to impute genetic variation in greater depth into the UK Biobank.
3 November 2023
Simon gave an invited talk on recombination and its evolution as drivers of genetic diversity in mammals at ASHG 2023.
9 August 2023
Sile’s preprint on modelling fine-scale population structure in the UK Biobank, on which Lino and Sinan are co-authors, is now available on bioRxiv. Sile et al. show that genetic effect sizes are conserved between human populations across a range of quantitative human traits.
11 July 2023
Ana’s new preprint on the distribution of branch duration and the detection of inversions in ancestral recombination graphs is out on bioRxiv.
17 March 2023
Luis, Miriam and Sinan recently passed their thesis defence vivas. Many congratulations, Drs Torada Aguilella, Stricker and Shi!
11 March 2023
Ana and Simon gave talks at the Probabilistic Modeling in Genomics Conference (ProbGen23) at the Cold Spring Harbor Laboratory in New York! Ipsita, Hrushi and Lino presented posters of their work.
28 October 2022
We recently welcomed two new members to the group: Dr Martin Houlard, who joins us as a senior postdoctoral researcher, and Isobel Myers, a new DPhil student co-supervised by Simon and Prof. Robert Davies.
25 March 2022
Emmanuelle is moving on to an exciting new job as Principal Scientist at Ochre Bio. We’re sad to see her go but wish her every success in her new role!
14 January 2022
We are looking forward to the 7th Probabilistic Modeling in Genomics Conference (ProbGen22), which will be held in Oxford (and also virtually in a hybrid format) from 28-30th March 2022. Simon and Robbie are part of the conference’s organising committee.
9 January 2022
We’re excited to welcome a new postdoctoral researcher, Ipsita Agarwal, into the group! Ipsita recently completed her PhD at Columbia University in New York City.
12 October 2021
Chris Gill, a DPhil student in the group, has passed his DPhil viva. Many congratulations, Chris!
11 October 2021
Anastasia Ignatieva has joined the group as a postdoctoral researcher. She has recently completed a PhD on the Oxford-Warwick Statistics Programme and will be working on developing scalable methods for understanding aspects of human evolution. Welcome, Anastasia!
We are also delighted to welcome Nik Baya, Yunong Lin and Hrushikesh Loya, three new DPhil students in the group!
27 September 2021
We are excited to welcome Brynelle Myers, a new research assistant in the group, and Jessica Cho, an undergraduate student joining us for a year-long placement. They will strengthen the group’s wet lab-based research team.
16 June 2021
Leo’s paper (with Robbie, Simon and others) titled “Inferring Population Histories for Ancient Genomes Using Genome-Wide Genealogies” is now published in Molecular Biology and Evolution! The paper introduces the Colate software for inferring coalescence rates for unphased, low-coverage genomes. This software is available for free on GitHub under an open-source license.
7 June 2021
“Rapid genotype imputation from sequence with reference panels”, a new paper by Robbie, Sinan, Simon and co-authors, has been published in Nature Genetics. QUILT, the algorithm for genotype imputation using low-coverage WGS data introduced in the paper, is freely available on GitHub under a GPL license.
17 February 2021
The pre-print of a new paper by Leo, Robbie, Simon and co-authors is now available on bioRxiv! The paper introduces an extension of the Relate algorithm to incorporate ancient genomes and infer their genealogies.
7th June 2020
Simon has been awarded the Weldon Memorial Prize! The prize is given yearly by the University of Oxford to the person who “in the ten years next preceding the date of the award, published the most noteworthy contribution to the development of mathematical or statistical methods applied to problems in Biology”.
30th October 2019
Our paper on ZCWPW1 is now avaliable on bioRxiv https://doi.org/10.1101/821678
2nd September 2019
Leo’s paper on genome-wide genealogy estimation for thousands of samples is now published in Nature Genetics.
Also see here for the corresponding news and views article by Kelley Harris!
25th June 2019
Daniel’s paper on single cell RNAseq in mice testis is now published in eLife.
17th June 2019
Simon is giving a talk at ESHG: “Genetic diversity and its unexpected impacts on recombination, genome evolution, speciation and sterility in mammals”
31st May 2019
Congratulations to Robbie, who completed his DPhil in our group, for his appointment as Associate Professor at the University of Oxford! Announcement on Twitter.
23rd May 2019
Mike’s paper on Fine-Scale Inference of Ancestry Segments Without Prior Knowledge of Admixing Groups was published in Genetics.
1st May 2019
Applications are invited for a full-time Postdoctoral Research Assistant in Statistical and Population Genetics, funded by Wellcome. Reporting to Professor Simon Myers and Dr Garrett Hellenthal, the post is fixed-term until 6 October 2020, there is a possibility of extension if appropriate. It will involve working within a collaborative group of leading researchers based in Oxford, UCL, and elsewhere.
Closing date: 10th June 2019, Full details: here.
14th February 2019
Our paper on genome-wide genealogy estimation is now avaliable on bioRxiv. https://doi.org/10.1101/550558
4th Feburary 2019
Clare’s paper on “Patterns of genetic differentiation and the footprints of historical migrations in the Iberian Peninsula” was published in Nature Communications.
A press release for this study can be read here.
4 - 7th November 2018
Leo and Daniel both gave talks at the ProbGen Conference in CSHL on “Reconstructing the genealogies of humans and wild mice” and “Decomposition of mouse spermatogenesis at single-cell resolution reveals dynamic transcriptional programs orchestrated by a rich regulatory repertoire”.
30th October 2018
Our paper on non-crossover events is now avaliable on bioRxiv. https://doi.org/10.1101/428987
12th September 2018
Simon will be giving a keynote presentation at the The Students and Postdocs Meiosis Workshop in Montpellier, France on the 12th of September.
22nd August 2018
Postdoc opportunity in statistical and population genetics at University of Oxford.
Closing date: 17th September 2018
We are currently advertising for a postdoctoral researcher in Statistical and Population Genetics at the University of Oxford’s Department of Statistics. Supervisory support will be jointly provided by Simon Myers and Jonathan Marchini (Oxford) and Dr Garrett Hellenthal (UCL). The post is for 18 months. For informal queries contact Simon Myers (myers@stats.ox.ac.uk). Full details at http://www.stats.ox.ac.uk/vacancies/.
16th August 2018
Daniel’s paper on single cell RNAseq of the testis is now avaliable on bioRxiv. https://doi.org/10.1101/393769
25th July 2018
Mike’s paper on inferring ancestry segments and characterizing admixture events is now avaliable on bioRxiv. https://doi.org/10.1101/376137
12th July 2018
Leo gave a talk at SMBE on “Identifying natural selection by constructing genome-wide genealogies”
12th March 2018
Clare’s paper on fine-scale population structure in Spain is now avaliable on bioRxiv doi.org/10.1101/250191.
6th February 2018
Clare and Simon have been working with Oxford Sparks to create an animation about their work on fine scale population structure as part of a special exhibition at the University of Oxford Museum of Natural History. You can watch the animation at youtube.com/watch?v=G6bNXdwEA84 and find out more about the Settlers exhbition at oum.ox.ac.uk/settlers/.
29th January 2018
Wellcome to Chris Cole who is joining the group for a rotation project as part of the Genomic Medicine and Statistics DPhil course.
3 Jan 2018
A number of our group are going to the 51st Population Genetics Group Meeting this week in Bristol. Leo will be giving a presentation on “A method for estimating genome-wide genealogies for thousands of samples”, and Jeffery will be presenting his previous post-doc work: “A numerical solution of the Wright-Fisher SDE with application to transition probability density approximation”.
15 December 2017
We went out for lunch this week to mark the departure of Ran who is off to do a second postdoc in the Ratcliffe group, Oxford.
06 December 2017

Simon delivered the Francis Crick lecture at the Royal Society on “The birth of a new species: Bringing together, yet driving apart”. (more details). If you missed it you can watch it on the Royal Societies YouTube channel here!
Image: Simon receiving his commemorative scroll from Sir John Skehel. (Daniel Wells CC-BY 4.0)
27 November 2017
Congratulations to Clare for passing her PhD viva this week!
26 October 2017
Our paper on “A map of human PRDM9 binding provides evidence for novel behaviors of PRDM9 and other zinc-finger proteins in meiosis” is now published in Elife.
27 August 2017
This week a number of us will be at the EMBO conference on Meiosis in Croatia.
31 May 2017
Nick’s paper “Human PRDM9 Can Bind And Activate Promoters, And Other Zinc-Finger Proteins Associate With Reduced Recombination In cis” is now available on bioRxiv!
18 July 2016.
Breaking news is that Simon has been awarded the 2017 Francis Crick Medal and Lecture by the Royal Society “for transforming our understanding of meiotic recombination and of human population history. ” The prize lecture will be delivered in December 2017.
This prestigious prize is a great recognition of the contribution that Simon is making to understanding key aspects of genetics.
04 July 2016.
Robbie and Simon (along with coauthors) have published a Nature Genetics paper detailing the STITCH method for Rapid genotype imputation from sequence without reference panels. This method allows for imputation based only on sequencing read data, without requiring additional reference panels or array data. The approach is accurate even in settings of extremely low sequencing coverage. For example, accurate imputation of 5.7 million SNPs at a mean r2 value of 0.98 in 2,073 outbred laboratory mice is achieved (0.15× sequencing coverage). In a related paper, also in Nature Genetics today, STITCH is used to do Genome-wide association of multiple complex traits in outbred mice.
15 April 2016.
We are hiring! There are 2 positions in Statistical Genetics Methods Development and 1 position in Statistical Genetics Software Development. The full-time positions are funded by the Wellcome Trust and are fixed-term for 3 years. These positions will focus on analysing data gathered by Genomics England (GEL). GEL will deep sequence the complete genomes of approximately 70,000 individuals, sampled across diverse rare disease phenotypes, cancer patients, and healthy individuals. Funding is via a Wellcome Trust Collaborative Award, and these positions will be supervised jointly by Prof Myers and Prof Jonathan Marchini.
The closing date for applications is 1 June 2016.
23 February 2016.
The 2nd Probabilistic Modeling in Genomics Conference (ProbGen16), will be held at the Department of Statistics, University of Oxford, UK.
Monday 12th September – Wednesday 14th September 2016
Organisers: Jonathan Marchini, University of Oxford, UK; Simon Myers, University of Oxford, UK; Jotun Hein, University of Oxford, UK; Peter Ralph, University of South California, USA
11 February 2016.
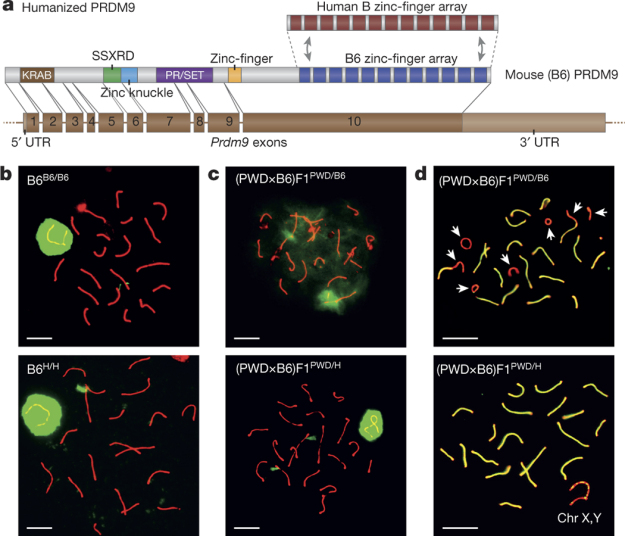
A study by a team including 5 members of the Myers Group has published a paper in Nature that shows the role the PRDM9 gene plays in speciation. This was achieved by completely restoring fertility in normally infertile hybrids of two mouse subspecies, by the insertion of a human equivalent of the PRDM9 gene.
16 November 2015.
Robbie is writing up his DPhil thesis and has found employment! He will be working with Genomics plc in Oxford as a Statistical Bioinformatician / Geneticist.
15 July 2015.

We had burgers and a few beers to mark the departure of Nick this week. Nick is off to do a second PhD in Bioengineering at UC Berkeley.
March 2015.
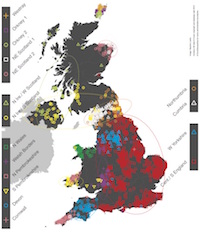
The People of the British Isles paper appeared in Nature. This paper leverages some of the recent work done by the Myers group and “shows that subtle variations in the genetic make-up of modern Britons reflect the history of their islands’ colonisation by peoples from mainland Europe since the last Ice Age.” (credit: Wellcome Trust Centre for Human Genetics)
February 2014.
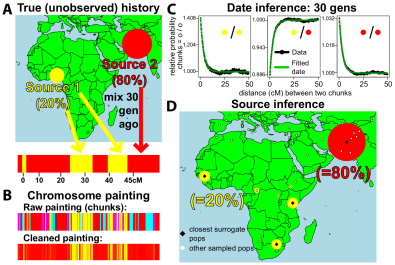
“A genetic atlas of human admixture history” appeared in Science. In this study, which attracted considerable press attention, we developed and applied to a large dataset of 1500 individuals from 94 populations, a model-based approach to identify, and date, mixing events among human populations. The work is the first to describe, in detail, how the DNA of most humans is affected by such events, and discovers impacts on our DNA of recorded historical events, e.g. the Mongol empire.